Easy Ways to Create Variations of Function
You will often need to create more than one version of a function. Here are two different methods, a piecewise function and using an Option. Either are fine, but in general simply for conciseness I use a piecewise function when the function is short (like less than a dozen lines) and an Option when the function is longer.Here is the dataset for the examples. To see the 6 functions used to create arrays in Mathematica, see Ways to Create Arrays.
dataset=Array[List,{5,3}]
{{{1,1},{1,2},{1,3}},{{2,1},{2,2},{2,3}},{{3,1},{3,2},{3,3}},{{4,1},{4,2},{4,3}},{{5,1},{5,2},{5,3}}}
Here's the first version. It takes a List of data as a first argument and an Integer as its second argument. This simple example uses the index to pick out a Part from the data.
function1[data_List,index_Integer]:=data[[index]]
function1[dataset,4]
{{4,1},{4,2},{4,3}}
The Piecewise Approach
Now we need a second version to take a List of indices in the second argument. We can simply use datatyping by specifying the Head of the second argument. This is one way to do the "piecewise" method. The domain of possible inputs is split into pieces (sub-domains) and each variation of a function is designed to pick out the correct piece (sub-domain) for which it is designed.function2[data_List,index_List]:=data[[index]]
function2[dataset,{2,4}]
{{{2,1},{2,2},{2,3}},{{4,1},{4,2},{4,3}}}
The Options Approach
You can see that if there were a minor change in a long function, copying the function is not as concise as the following approach using Options. There is more overhead in this example to create the Option and handle it in the function, but in a long function that overhead is less than copying as in the Piecewise approach.Note that for concise creation of Piecewise mathematical functions, the built-in function Piecewise should be used. For an explanation of how I use Options, see A Template for Optional Arguments [to be published].
ClearAll@function3;
Options@function3="indexOption"->"Integer";
function3[data_List,index_,options___?OptionQ]:=
Module[{indexOption="indexOption"/.{options}/.Options@function3},
If[indexOption=="Integer",dataset[[index]] ];
If[indexOption=="List",dataset[[{index}]] ];
(*endModule*)]
Here is the function that prompted me to write this primer. It takes the membrane voltage time series from simulated spiking neuron cells and counts how many spikes occur, with cell range and time series range as arguments. I needed to expand it so I could specify counting spikes in several time series ranges. I didn't want to copy and vary it with the piecewise approach, so I used the options approach. The changes to the original function to implement the new ones are italicized.
ClearAll@batchMeanFiringRateTable;
Options@batchMeanFiringRateTable={"Export"->Off,"MultipleBatch"->Off,"MultipleTimeSeries"->False};
batchMeanFiringRateTable[spikeFileDirectory_String,cellRange_List,timeSeriesRange_List,leftColumnHeading_String:"Neural Group",rightColumnHeading_String:"Average Firing Rate",options___?OptionQ]:=
Module[{exportFileName,firingData,
spikeFiles=getSpikeFiles@spikeFileDirectory,
exportOption="Export"/.{options}/.Options@batchMeanFiringRateTable,
multipleBatchOption="MultipleBatch"/.{options}/.Options@batchMeanFiringRateTable,
multipleTimeSeriesOption="MultipleTimeSeries"/.{options}/.Options@batchMeanFiringRateTable},
(*With "Batch"\[Rule]On, meanFiringRateFromSpikeFile will just return the averageSpikeRate each time it's called*)
(*Need FileBaseName to identify cell group in left column if TableForm; need First@timeSeriesRange to remove the outer List for a single time series range*)
If[multipleTimeSeriesOption==False,firingData=Table[{FileBaseName@batchFile,meanFiringRateFromSpikeFile[batchFile,cellRange,First@timeSeriesRange,"Batch"->On]},{batchFile,spikeFiles}] (*endIf*)];
If [multipleTimeSeriesOption==True,firingData=Table[{FileBaseName@batchFile,meanFiringRateFromSpikeFile[batchFile,cellRange,timeSeries,"Batch"->On]},{batchFile,spikeFiles},{timeSeries,timeSeriesRange}]/.{{x_,y_},{x_,z_}}->{x,y,z};(*Table will take each spike file and iterate over timeSeriesRange. Don't need First@timeSeriesRange here. *) (*endIf*)];
If[multipleBatchOption==Off,Print@"Multiple batch is off.";Print[firingData//TableForm[#,TableAlignments->Left,TableHeadings->{None,{leftColumnHeading,"Ave AP Rate: "<>ToString@timeSeriesRange}}]&],Return@firingData (*endIf*)];
If[exportOption==On&&multipleBatchOption==Off,exportFileName=FileNameJoin@{DirectoryName@spikeFiles[[1]],ToString@FileNameTake[DirectoryName@spikeFiles[[1]],-1]<>"-BatchFileTable.xlsx"};
Export[exportFileName,firingData];Print@StringForm["File exported to ``.",exportFileName]];
]
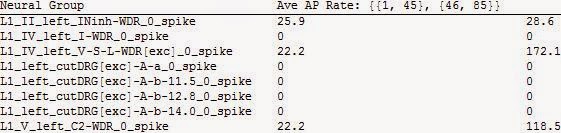
batchMeanFiringRateTable["C:\\Users\\Public\\Documents\\UNCuS16_09_2013\\DataFiles\\WDR-Abeta-14-12only\\WDR-Abeta-14-12only-CS8-9-20-RS0p28\\",{1,80},{{1,45},{46,85}},"Neural Group","Method"->"OverSpikingCells","MultipleBatch"->Off,"Export"->Off,"MultipleTimeSeries"->True]

No comments:
Post a Comment